William Harms
RTOG 3D Quality Assurance Center
Washington University
510 South Kingshighway Blvd.
St. Louis, MO 63110
Version 4.00
12 October 1999
Based on AAPM Report #10 and as used and modified by the NCI Particle Intercomparison Contract, the NCI High Energy Photon External Beam Treatment Planning Contract, the NCI Electron External Beam Treatment Planning Contract, and the RTOG 3D QA Center.
This Tape/Network Format Specification, while initially based on AAPM Report #10, has been significantly altered to allow more information to be included in the data transfer. It was originally modified by the NCI Particle Intercomparison contract, then used in that form by the NCI High Energy Photon External Beam contract. The document was modified further for the NCI Electron External Beam contract. The modification in this version reflect further trimming of unused image types with the intent to add more image types that directly impact on exchange of treatment planning and treatment verification.
A significant modification was made with version 3.00 as it included several heretofore unsupported data image types. These new image types include beam geometries, digital film images, and dose-volume histograms. Additionally, several changes were made to dose distributions to remove ambiguities involving the submission of other than absolute dose.
With Version 3.10, an apparently ambiguous keyword was removed and more clarifying comments and examples were added. An additional keyword was added for beam geometry to identify the algorithm used for calculating doses from the beam. All of these additional keyword additions, or deletions, are optional in nature to maintain compatibility with Version 3.00. To simplify network exchange of these data files, the requirement for "buffered" data blocks is removed as an option (to be agreed upon by sending and receiving site). As many institutions are originally writing their files in this format and then post processing them to "block" them, this should come as a welcome change. This "unbuffered" submission is only available for electronic exchange of the data and the use of buffers will still be required for any tape media data exchanges.
Version 3.20 added an additional keyword for digital film images "Collimator Angle" (primarily DRR's without portal marking in the image) and one for beam geometry "Head In/Out". These were to clear up ambiguities and oversights in the previous version. DRR's are computed by two primary geometric methods, one removes the collimator angle from the transformation matrix used for computing the DRR and the other always has the edges of the image parallel to the collimators (the collimator angle is left in the transformation matrix). The "Collimator Angle" keyword identifies the method being used and is optional if the DRR edges are parallel to the unrotated collimator. The "Head In/Out" was added to resolve potential ambiguity in the couch angle wherein a 180 degree offset was added to the couch angle to signify a foot in treatment. If that patient is being treated with their feet to the gantry (prior to any couch rotation), this keyword must be used, otherwise, head in is assumed.
Changes were made in the document for Version 3.21 which mostly amounted to additional explanation of keywords and data inclusion. There were several keywords for Beam Geometry, Digital Films and Dose-Volume Histograms which were moved from the Required Keywords to Optional Keywords. This was primarily to simplify the directories by removing requirement on any data which was not necessary to interpreting the data provided in the file set.
As more institutions begin to participate in studies requiring the use of this data exchange specification, it is inevitable that further refinement and ambiguity resolution will have to be done. This is a living document and will be subject to many revisions over the next year or two until it is replaced with more robust and universal communication mechanisms such as DICOM 3.0.
Version 4.00 was created to provide for ultrasound guided permanent prostate seed implants. Additional items were added in support of Peregrine and other projects. Those which were added which are not supported by the 3D QA Center are identified as such in the text.
| Version | Date | Description |
|---|---|---|
| 1.0 | 4/22/82 | Preliminary draft. (Michael Goitein) |
| 1.1 | 8/28/82 | Substantially modified. All images in ACSII except CT scans |
| 1.2 | 10/21/83 | Intermediate update - never distributed. |
| 2.1 | 12/27/83 | Working version. Document clarified and reorganized. New requirement that CT images be contiguous on tape in order of increasing z-coordinate Explicit description of how null characters are to be handled. (nulls not included in byte counts). |
| 2.2 | 4/08/85 | Revisions made in conjunction with Robert F. Curley: Add dose examples
Add text describing in words the data files for structures and doses. Require
"....." must not contain CR/LF Require all CT scans to be square. Add a number of clarifying comments. |
| 2.3 | 09/22/89 | Remove annotations and code examples for ECWG report (Harms) |
| 2.4 | 07/08/92 | Remove additional information on annotations, cleaned up the grammar, added variances relating to the amount of data on a tape (multiple patients, buffer size, and tape density), added new "image" types of MRI, Beam Geometry, and digital film images (i.e. DRR, on-line images). (Harms) |
| 2.5 | 06/29/93 | Fixed errors in document pertaining to keywords "Maximum # of scans" to "Maximum # scans" and "Scan type" to "Scanner type" (Harms) |
| 2.51 | 07/09/93 | Clean up language and add "Writer" as a Directory Header entry (as it was inadvertantly left out from the original format) |
| 3.00 | 1/10/94 | Added Beam Geometry, Digital Film, DVH's and fractionation information. Included moving appendices into appropriate chapters and modified dose distributions to allow for fractionation information and to clarify dose units. (Prostate Working Group, Bill Harms, Jonathan Jacky, Jeff Lewis and James Balter). |
| 3.10 | 6/10/94 | Added more explanation and cleaned up some partial omissions. Allowed unblocked data (for network transmission) if receiving site is agreeable and removed "INTERCOMPARISON STANDARD #" as an ambiguous keyword. Removed AAPM Report 10 as the standard to judge discrepancies in the exchange format. |
| 3.20 | 12/28/94 | Added "Head In/Out" keyword to beam geometry and "Collimator Angle" keyword to digital film images for DRR's. |
| 3.21 | 3/8/95 | Added clarifying discussion to many keywords and moved unnecessary keywords from Required, to Optional. |
| 3.22 | 4/17/97 | Corrected error in MLC example. |
| 3.30 |
7/25/97 | Optional extensions added to Beam Geometry and Dose for support of Peregrine
communications and more succinct and explicit treatment plan information exchange. Some
additional clarification text was incorporated based on comments by George Starckschall.
The primary additions to Beam Geometry are explicit compensating filter descriptions, a
Machine ID keyword (in addition to energy and modality), and Beam Weight and Weight Units
to allow for machine settings to be specified. The optional additions to Dose allows for
binary dose files (two's complement integer with a scale factor) to reduce the size of
dose image files. Following are links to the modified text for ease of locating.
|
| 4.00 Draft |
03/22/1999 | Changes made for Version 4.0 of this Exchange Specification were
motivated by the need to add prostate seed brachytherapy treatment planning to the
information supported by this exchange. In order to make use of the appropriate
imaging modalities which are used for permanent prostate seed implants, MRI and ultrasound
(US) were added to the CT Scans chapter and an additional file type was defined for Seed
Plan specification. One previously documented feature has been removed. While the Specification "officially" supported multiple patient data sets in a single file set, no commercial or University systems being used for patients enrolled in multi-institutional trials made use of this feature, therefore, to simplify the implementation of reading and writing software, this feature has been removed. This allows the Case keyword to be used as desired by the writing facility. A suggestion would be to use the actual patient registration number as the case number, but in order to maintain backward compatibility with writing software, this is not going to become a requirement at this time. One additional change was to incorporate beam aperture definitions through the use of a transmission table in addition to closed block and portal contours. This addition was made to facilitate the exchange of this information with the Peregrine system and is not currently used, or supported by the RTOG 3D QA Center for protocol patient data submissions. Following are links to the modified text for ease of locating. Also, note that all added text is in this same color purple text to aid the reader. Incorrect compensating filter examples were also corrected.
|
| 4.00 |
10/12/1999 | Changes made from the Draft Version 4.0 of this Exchange Specification
were the result of the implementation workshop in September, 1999 where most of the
brachytherapy developers provided input into items requiring correction or adjustment.
These changes generally added clarification to the document, but in some cases,
removed some of the draft items or added items which would not result in legacy code
failing. The optional keyword for Seed Geometry, "Registered" has been removed and replaced with the implicit assumption that if any CT/MR/US images are included in the same file set as one (or more) Seed Geometry files, the seeds are registered with the included images. Following are links to the modified text for ease of locating. Also, note that all added text is in this same color purple text to aid the reader.
|
The format proposed follows the recommendations of the AAPM for digital image transfer, published as AAPM report no. 10, "A Standard Format for Digital Image Exchange" (obtainable from: AAPM, One Physics Ellipse, College Park, MD 20740). The description in this document assumes the reader's familiarity with AAPM Report #10. The tape format described in this document is intended to comply with all aspects of AAPM Report #10. Some aspects of that report are reiterated here as a help to the reader. However, in the event of a real or apparent discrepancy, AAPM Report #10 shall give way to this document. This document extends the scope of AAPM report #10 by including data structures other than CT scans or comparable images.
Seven types of files (termed images in the AAPM standard) are supported (in addition to the Directory): Comments; CT scans; Structures (target volumes, external contours, normal critical structures, etc.); Beam Geometry's; Dose distributions; Digital Film Images and Dose-Volume Histograms. No more than one case can be transmitted on one tape (or network file data set). The data shall be placed on tape in the following order, case by case.
| Comments | case 1 |
| Scans (CT, MRI, US) | case 1 |
| Structures | case 1 |
| Orphan Digital Film Images | case 1 |
| Beam Geometry's | case 1 (plan 1) |
| Digital Film Images | case 1 (plan 1) |
| Doses | case 1 (plan 1) |
| Dose-Volume Hist. | case 1 (plan 1) |
| Beam Geometry's | case 1 (plan 2) |
| Digital Film Images | case 1 (plan 2) |
| Doses | case 1 (plan 2) |
| Dose-Volume Hist. | case 1 (plan 2) |
| ......................... | ................. |
| Beam Geometry's | case 1 (plan n) |
| Digital Film Images | case 1 (plan n) |
| Doses | case 1 (plan n) |
| Dose-Volume Hist. | case 1 (plan n) |
| etc. |
Not all data is required in this order. For instance, if beam geometry's and digital film images are not submitted with the corresponding doses and dose-volume histograms, the non-existent data will just be left out of the data to be transmitted. An example of such an order would be:
| Scans | case 1 |
| Structures | case 1 |
| Doses | case 1 (plan 1) |
| Dose-Volume Hist. | case 1 (plan 1) |
| Doses | case 1 (plan 2) |
| Dose-Volume Hist. | case 1 (plan 2) |
| ......................... | ................. |
| Doses | case 1 (plan n) |
| Dose-Volume Hist. | case 1 (plan n) |
| etc. |
Seed Geometry and Beam Geometry are mutually exclusive and both may not be contained in a single file set. In the case of a file set containing Seed Geometry, the following demonstrates the order of the files:
| Scans (CT/MR/US) | case 1 |
| Structures | case 1 |
| Seed Geometry (one set of seeds) | case 1 (plan 1) |
| Doses | case 1 (plan 1) |
| Dose-Volume Histograms | case 1 (plan 1) |
| Seed Geometry (second set of seeds in same implant) | case 1 (plan 2) |
| Doses | case 1 (plan 2) |
| Dose-Volume Histograms | case 1 (plan 2) |
| etc. |
If multiple Seed Geometry files are contained within a given file set, it is assumed that they represent different activity and/or model seeds used in the same implant.
Examples of a directory header and some (non-binary) images are included in the following chapters.
There are two distinct coordinate systems used by this Specification. One is for patient data which is defined in Chapter 6. The other is for the beam aperture specification which is oriented in a "beam's-eye view" manner in which aperture coordinates are 2D coordinates with a constant third coordinate relative to distance from beam source and is defined in Chapter 8.
A 9-track tape with a density of 1600 bpi shall be the default medium used for data exchange. However, if the site to receive the tape agrees to higher density, and/or a different type of physical tape, it shall be allowed. Tapes shall be UNLABELED to facilitate intercommunication between different manufacturer's computers. Multi-volume tapes should not be used unless necessary to transmit a single case. For tapes which can have their densities changed, the tape must be clearly labeled and the used density agreed to by the receiving institution.
All data on the tape shall be written in fixed length buffers. The default buffer size is 2048 bytes, but if the receiving site agrees to a different size buffer, it is allowed and should be clearly marked on the tape. As many buffers are written as are required to transmit the data, unused bytes (such as the unused remaining bytes of the last buffer of an image) shall be filled with NULL characters. No text strings should be broken across buffer boundaries. If an entire string will not fit into the current buffer, the end of the buffer should be NULL'ed out and the string put into the next buffer.
Single end-of-file marks separate the directory file from the first "image" file and succeeding image files from one another. Two end-of-file marks in succession terminate the tape. On media other than 9-track tape, these separation requirements may not be valid and adjustments may need to be made.
If both the sending and receiving site have network access to one another, this data may be sent as individual files across the network. The means of such transfer are left for the sending and receiving institutions to work out among themselves. Recent experience has shown that anonymous ftp, in binary mode, is a practical method of such data transfer where the files' names have a numeric identifier in their names so that the order is obvious for processing (the author's preference is "aapm0000", "aapm0001", etc.). However, anonymous ftp might present patient record confidentiality problems. This could require the submitting institution(s) to have distinct login accounts on the receiving machine(s) which segregate them from other institutions data submissions and shield the data they submit from others.
For network exchange of data, if the receiving site agrees, the data may be sent in files of a single buffer the size of the data file. The fixed length buffer requirement may be disregarded in this case. However, for media exchange of data, in the interest of preventing any possible hardware/software incompatibility, fixed buffers are still REQUIRED. This is a change for Version 3.10.
Two types of data can be stored on tape: BINARY data, for CT scans and digital film images; and ASCII character strings (terminated with <CR/LF>) for everything else (including the directory file). The two types of data may NOT be mixed within any given file.
For each binary datum which occupies 2 bytes of the buffer, in compliance with the AAPM standard, the most significant byte is required to be first. Thus VMS, and similar byte order, machines will need to byte-swap both when writing and when reading a tape, for instance. For the unsigned byte data, the order is obvious.
ASCII data may appear in one of two contexts: In the directory header where the data is always in the form of keyword/value pairs (see below); and in images (such as structure definitions or dose distributions) - where the format depends on the data type, but is generally largely a sequence of numeric fields (i.e. ASCII strings defining real or integer numbers as appropriate). In either context the following rules apply.
Each entry of ASCII text may be from 1 to 80 bytes in length (excluding null bytes which are ignored) and must be terminated by a carriage-return/line-feed (CR/LF) sequence (not included in the 80 byte limit). Embedded spaces, tabs and null characters should not be included within numeric fields (but may precede or follow them) and elsewhere (as in keywords) they are to be ignored. Blank lines (CR/LF/CR/LF) are to be ignored in the parsing of these files. To permit comments in numeric fields (in order to make a printed file more interpretable), any text enclosed in double quotation marks (") is to be ignored. Text between quotation marks may not include a CR/LF string.
When specifying numeric data, a comma/space (comma followed by a space) sequence is an acceptable field delimiter as well as the CR/LF sequence. ADD1: Note, however, that no text line may end with a comma/space/CR/LF sequence as the comma/space implies further meaningful text in the line. No text string may bridge multiple buffers, if buffered exchange is selected or required. While the specification technically allows it, it generally presents implementation problems and shall not be supported.
Unused elements of the last buffer of a binary image (if any) are ignored. They may be filled with zeros.
Null characters may occur anywhere within ASCII Text (except in the middle of a numeric field) and are to be ignored. Null characters are not counted in any per line byte count limit. Generally, it is expected that null characters will be used to pad out at least the final buffer of an image, and should be used to pad out the final elements of intermediate buffers to avoid having text cross buffer boundaries. Only binary data may cross buffer boundaries.
The first file is a directory file, written entirely in ASCII characters. The directory consists entirely of Keyword/Value pairs - as described in the AAPM standard specification and in this document. At present no files or "images" other than the directory contain keyword/value sequences. Keywords and values are case and space insensitive. For instance:
Somewhat longer keyword :=
is equivalent to:
SOMEWHAT LONGER KeywoRd :=.
The first entries in the directory pertain to the entire tape and constitute the "directory header". Keywords used in the directory header are given in the following section. The directory header is followed by sequences of keywords which relate to individual images. By convention the first such keyword shall be "Image #", and all keywords relating to an image should follow that "Image #" specification and should precede the next "Image #" occurrence.
Note that "Image #" is a misnomer introduced by the AAPM format for tape exchange. It really just identified the position of the file on the tape. However, it does reference the sequence number of the associated file for network transferred data files. The first file is the directory (perhaps best thought of as file number 0), and subsequent files are termed "images" and assigned consecutive numbers starting from 1. In the present case, these "images" may in fact be any one of: Comments, CT scans, Structures, Beam Geometry's, Digital Film Images, Dose Distributions and DVH's.
Spaces, tabs and null characters are to be ignored in keywords. Alphabetic characters may be in upper or lower case and, in interpreting strings of characters as keywords (program implementation), all lower case characters may be replaced by their upper (or lower) case equivalents. In order to remove a potential source of confusion, the character strings "number" and "#" in keywords are to be everywhere considered interchangeable and MUST have numeric values.
In conformity with the AAPM standard, directory entries are made in the format:
Keyword := value
In this context only one "value" can follow the "=". Thus a mixed expression such as "size : = 1.5 cm" is illegal. There is to be no character (null, space, or otherwise) between the ":" and the "=".
In order to make tape listings somewhat more readable, it is permissible (indeed encouraged) to include tabs to make successive entries line up, as:
Keyword := STRUCTURES
Somewhat longer keyword := 18
Next keyword := 10.65
The AAPM tape format virtually mandates a two-pass approach - that is, two passes have to be made through the data to be transferred: the first in order to build up and write out the entire directory; the second in order to write out the underlying data to tape. This may be avoided if the files are built on disk first and the physical writing of the tape subsequent to the completion of all data files and the directory being written to disk. Network transfer will involve building the files on disk with the directory file being written to disk last (even though it has a smaller file number, i.e. 0).
Required Keywords
Tape standard # := 4.00 (version # of this standard from title page)
Institution := Name of submitting institution
Date created := Date tape written in AAPM format (dd, mm, yyyy)
Writer := Name of person responsible for writing tape
These entries must be the first entries in the directory.
Optional Keywords
Intercomparison standard # := version # of this standard from title
page (4.00) this keyword is maintained
only for compatibility and its' use is
not recommended
Format of data in the image:
No image is associated with the directory header.
Tape standard # := 4.00
Institution := MIR
Date created := 22, 03, 1999
Writer := Bill Harms
The date format used for all dates specified in a directory for a data exchange file set must be in the format DD, MM, YY[YY], where DD is the day of the month (one or two digits are allowed), MM is the month of the year (one or two digits are allowed and 1-January, 2-February, etc.), and YY is the last two digits of the year with an implied 1900 added to it. Four digits may be used for the year for Y2K compliance (and must be used after 12/31/1999).
Note that a date may be legal in format, but due to the time of any given month in which the date is generated, it may be incorrect. For instance, if a file set is generated on the 9th of February 1995, the date string should be 9, 2, 95. However, 2, 9, 1995 is a legitimately formatted date, but is incorrect. This should be carefully reviewed during implementation as it is a frequent mistake.
There are four keywords which are common to all image files (regardless of the image file content). These keywords must be used for all image files and must be in the order specified for the proper implementation of the data exchange format.
Required Keywords
Image # := actual image (file) number
Image type := COMMENT, CT SCAN, MRI, ULTRASOUND, STRUCTURE,
BEAM GEOMETRY, DIGITAL FILM, DOSE,
SEED GEOMETRY, or DOSE VOLUME HISTOGRAM
Case # := 1 for first case(optionally protocol case #)
Patient name := patient identifier
The Image # is the ordinal number of the data file being referenced. In the case of tape being used as the transport medium, this number is the order in which the files are found on the tape in which the first file is the directory file and is considered file number zero (0). Therefore the first data file would be 1, the second, 2, etc. In the case of a network medium of exchange, these number must be explicitly represented in the file names attached to the individual files. Again, the directory file is file zero (0).
The Image type is used to identify the data contained in the associated image file. With the exception of CT SCAN, MRI, ULTRASOUND, DIGITAL FILM and binary dose files (optional) all data files are in ASCII format.
The Case # identifies the ordinal value of a patient in an exchange file set. Since multiple patient data sets are eliminated from this specification, this number may have any integral value and one suggestion would be to make it represent the case number assigned by the cooperative group for the protocol the patient is enrolled in.
The Patient name is not required to be the patient's real name. However, it should have the same value for all image files for the same patient in the exchange file set. For RTOG 3D QA Center purposes, it should be the patient's name or some other identifier which the submitting institution can use to identify the data set in question should the 3D QA Center have questions about the case..
Image # := 1
Image type := COMMENT, CT SCAN, STRUCTURE, BEAM GEOMETRY,
DIGITAL FILM, DOSE, or DOSE VOLUME HISTOGRAM
Case # := 1
Patient name := John Q. Public
This feature provides the capability of transmitting substantial textual material such as a clinical case history. The format of the data is as a sequence of ASCII text strings, each of no more than 80 characters, of arbitrary length. Although the comment text can be entered in any way desired, the most likely mechanism would be to provide a utility to read a file created with the computer's text editor and copy it into the comment "image" after adding the appropriate <cr/lf> line terminators and buffering. An example in 5.3 illustrates this.
Required Keywords
Image # := actual image (file) number (see 4.4)
Image type := COMMENT
Case # := 1 for first case, 2 for second case, etc.
Patient name := patient identifier
Optional Keywords
Writer := person responsible for data
Date written := date file written (DD, MM, YYYY)
Unit # := data identifier (submitting site)
File of origin := Name of original file.
Comment description := brief title to characterize comments
Format of data in the image:
ASCII text.
Image # := 1
Image type := COMMENT
Case # := 1
Patient name := FALSENAME
Unit # := 01-23-456
Comment description := Example of a comment file
This is an example of comment text. It can be used to transmit information about the case being transmitted, or anything else.
Many such "images" can be put on one tape, and more than one can apply to any one case. The directory entry "comment description" is a useful way of indicating what is in this file so that the recipient of the tape can decide on the urgency with which to approach the task of looking at the comment.
CT scans, MR images and ultrasound images (hereafter referred to as Patient Images or PI) are two dimensional arrays of 8 or 16 bit numbers. In the case of the 16 bit numbers, they are to be packed most significant byte first in accordance with the AAPM format. Patient Images (PI) are required to have square pixels (size of grid 1 units is the same as the grid 2 units). With the publication of Version 4.00, non-squarePI are now supported. The PI pixel numbers are required to be POSITIVE in the range 0 to 32767 for 16 bit pixels and 0 to 255 for 8 bit pixels - which means that some offset must be added to the Hounsfield (or other) numbers natural to the scanner to ensure that this constraint is complied with. In the case of 8 bit data, the data type is unsigned byte which requires that if the 8 bit data is handled as positive and negative values on the submitting system, an offset must be provided to ensure proper order of the pixel values..
To define the CT scale fully the user is required to provide two constants, CT-AIR and CT-WATER which are, respectively, the values of the transmitted data which correspond to air and water. If, for example, the user added 1000 to CT numbers of a scanner which has -1000 to +3071 as the normal range of CT numbers, the constants would have the values CT-AIR = 0 and CT-WATER = 1024 when the CT values are offset by +1000. The CT offset should be large enough that no negative binary values are written in the CT data and no CT value is greater than 32767.
A new keyword (IMAGE SOURCE) has been added to provide for descriptive information about the source of the CT/MR/US images. When this optional keyword is not used the source is assumed to be the image acquisition device (i.e. scanner, ultrasound unit, etc.). When this keyword is used, the only allowed value for the key value is SECONDARY CAPTURE. Using SECONDARY CAPTURE assumes that the images were acquired by some type of frame capture, either digitizer or screen capture. Several heretofore required keywords become optional when the IMAGE SOURCE keyword is used. The CT AIR and CT WATER values are no longer required when SECONDARY CAPTURE is the image source. Also, the pixels are allowed to be rectangular when SECONDARY CAPTURE is the image source.
Many scanners have an imperfect CT scale, so that air and water do not have their nominal values. This can be corrected by supplying the correct values (rather than the nominal values) for CT-AIR and CT-WATER. Non-linear behavior is possible. If the user has corrected for this the keyword/value "CT scale := Linearized" must be provided. If the CT numbers have been transformed to water-equivalent densities the keywords/value "CT Scale := Water-equivalent" must be provided. If the CT numbers transmitted should be distrusted above the certain value, that value should be specified with the "Distrust above" keyword.
The pixel data are to be ordered so that, if a scan is considered to be viewed from the patient's feet, the first pixel would correspond to the upper left hand corner of the scan, subsequent pixels would correspond to the data in the first row going from left to right followed by the pixels of the second and subsequent rows, ending at the lower right hand corner.
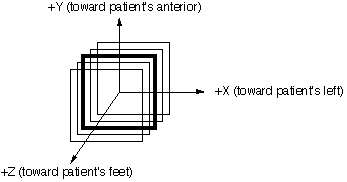
A right-handed cartesian coordinate system - referred to as the PATIENT COORDINATE SYSTEM - is superimposed on the scans. ADD2: The z axis is positive pointing out of the paper, which always points toward the patient's feet. It should be noted that this is DIFFERENT from the IEC patient coordinate system.

Figure 6.1
Figure 6.1 illustrates the coordinate system. The axes depicted in Figure 6.1 represent a patient who is scanned head first in a supine position. The coordinate system is more accurately described as a "hybrid" coordinate system where X and Y are independent upon the patient orientation on an external beam treatment unit couch and the Z coordinate is based on patient scan order. While the Figure 6.1 anatomical labels correspond to the identified axes when scanned head first, supine, the X and Y coordinate axes are actually tied to a treatment couch with +X to the right of the gantry when viewed from the couch and +Y is up toward the ceiling (assumes couch position orthogonal to plane of gantry rotation). The +Z coordinate is always toward the patient's feet independent of their scanning or treatment orientation which may require inverting this coordinate value depending upon the order maintained by the RTP system. With regard to coordinate system for brachytherapy data exchange, the anatomical labels and the corresponding axes identified in Figure 6.1 must be used.
Generally the origin of the patient coordinate system is at the dead center of the CT/MRI/US image (element 160.5, 160.5 of a 320x320 array, for instance where 1 refers to the first pixel in the image). However, offsets of the images are permitted as indicated in the following figure. Offsets are positive when the displacement is in the indicated directions as in Figure 6.2 (i.e. they are the directional measurement from the patient coordinate system origin in X and Y to the geometric center of the scan).
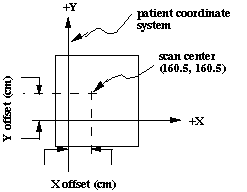
Figure 6.2
Scans must be provided in contiguous order on tape (or in the file set), in order of monotonically increasing value of the z coordinate. However, a sequence of scans need not be uniformly spaced along the axis normal to the plane of the scans (z axis).
In terms of this coordinate system, CT/MRI/US data are to be stored within the data array in the following manner: The upper left hand corner pixel (least x, greatest y) is first, followed by pixels in the first row (i.e. the x dimension is incremented first), followed by subsequent rows of lesser y value until the bottom right (greatest x, least y) pixel terminates the array. With the exception of some keyword changes, the MRI/US image format is almost identical to that of the CT scan images both in terms of the actual pixel data as well as in the directory structure entries.
Required Keywords
Image # := actual image (file) number (see 4.4)
Image type := CT SCAN identifies as CT scan
Case # := 1 for first case, 2 for second case, etc.
Patient name := patient identifier
Scan type := TRANSVERSE
CT offset := see text
Grid 1 units := pixel width (cm)
Grid 2 units := pixel height (cm) Must be same as Grid 1
units unless IMAGE SOURCE is used
Number representation := TWO'S COMPLEMENT INTEGER
Bytes per pixel := must equal 2
Number of dimensions := must equal 2
Size of dimension 1 := number of rows
Size of dimension 2 := number of columns
z value := couch position (cm, + to feet)
x offset := usually 0.0 (cm) [signed x distance from
coordinate system's x origin to the
geometric center of the CT scan pixel image]
y offset := usually 0.0 (cm) [signed y distance from
coordinate system's y origin to the
geometric center of the CT scan pixel image]
CT-air := 0 (this value is optional when the
IMAGE SOURCE is used)
CT-water := 1000 (this value is optional when the
IMAGE SOURCE is used)
Optional Keywords
Unit # := Unit number or ID
Site of Interest := prostate, etc. as appropriate
Scan description := "contrast study", etc.
Scanner type := GE9800, SIEMENS DRH, etc.
Head in/out := IN, OUT
Position in scan := NOSE UP, NOSE DOWN, LEFT SIDE DOWN,
RIGHT SIDE DOWN
Patient attitude := RECUMBENT, SEATED, STANDING
Tape of origin := helps you retrieve your original data
Study number of origin := helps you retrieve your original data
Scan ID := original scan identifier
Scan # := scan # in this sequence
Scan date := use AAPM format (DD, MM, YYYY)
Scan file name := original file name
Slice thickness := in cm.
CT scale := LINEARIZED, WATER-EQUIVALENT
Distrust above := maximum credible CT value
Image Source := SECONDARY CAPTURE
The pixel sizes (Grid 1 or 2 units) are positive for transverse oriented images. All coordinates and linear dimensions are expressed in centimeters.
Format of data in the image file:
Binary data in two's complement integer 0 to 32767.
Only the first two scans of this data set are shown.
Image # := 1
Image Type := CT SCAN
CASE # := 1
Patient name := BREAST1B
Scan type := TRANSVERSE
CT Offset := 1024
Grid 1 Units := 0.0938
Grid 2 Units := 0.0938
Number Representation := TWO'S COMPLEMENT INTEGER
Bytes per Pixel := 2
Number of Dimensions := 2
Size of Dimension 1 := 512
Size of Dimension 2 := 512
Z value := 7.5000
X Offset := 0.0000
Y Offset := 0.0000
CT-air := 0
CT-WATER := 1024
SCAN # := 1
Slice Thickness := 0.5000
Image # := 2
Image Type := CT SCAN
CASE # := 1
Patient name := BREAST1B
Scan type := TRANSVERSE
CT Offset := 1024
Grid 1 Units := 0.0938
Grid 2 Units := 0.0938
Number Representation := TWO'S COMPLEMENT INTEGER
Bytes per Pixel := 2
Number of Dimensions := 2
Size of Dimension 1 := 512
Size of Dimension 2 := 512
Z value := 8.0000
X Offset := 0.0000
Y Offset := 0.0000
CT-air := 0
CT-WATER := 1024
SCAN # := 2
Slice Thickness := 0.5000
and so on for the remainder of the scans.
Data are in 16-bit, 2's complement, integer representation but are required to be within the 0 to 32767 range. Data is in raster order with the first pixel being the upper left of the image (i.e. the most negative x coordinate pixel and the most positive y coordinate pixel), the next pixel being just to the right of the first pixel until that raster line is complete, then all remaining raster lines until the last pixel (lower right).
Required Keywords
Image # := actual image (file) number (see 4.4)
Image type := MRI or ULTRASOUND
Case # := 1 or Registered case number (numeric only)
Patient name := patient identifier
Scan type := TRANSVERSE
Pixel offset := value added to each pixel to ensure >= 0 for all pixels
Grid 1 units := pixel width (cm)
Grid 2 units := pixel height (cm) Must be same as Grid 1
units unless IMAGE SOURCE is used
Number representation := TWO'S COMPLEMENT INTEGER or UNSIGNED BYTE
Bytes per pixel := 2 for two's complement or 1 for unsigned byte
Number of dimensions := must equal 2
Size of dimension 1 := number of rows
Size of dimension 2 := number of columns
z value := couch position (cm, + to feet)
x offset := usually 0.0 (cm) [signed x distance from
coordinate system's x origin to the
geometric center of the CT scan pixel image]
y offset := usually 0.0 (cm) [signed y distance from
coordinate system's y origin to the
geometric center of the CT scan pixel image]
Optional Keywords
Scan date := use AAPM format (DD, MM, YYYY)
Image Source := SECONDARY CAPTURE
The pixel sizes (Grid 1 or 2 units) are positive for transverse oriented images. All coordinates and linear dimensions are expressed in centimeters.
Format of data in the image file:
Binary data in two's complement integer 0 to 32767 or byte 0 to 255.
Only the first two scans of this data set are shown.
Image # := 1
Image Type := MRI
CASE # := 1
Patient name := BREAST1B
Scan type := TRANSVERSE
Pixel Offset := 127
Grid 1 Units := 0.0938
Grid 2 Units := 0.0938
Number Representation := UNSIGNED BYTE
Bytes per Pixel := 1
Number of Dimensions := 2
Size of Dimension 1 := 256
Size of Dimension 2 := 256
Z value := 5.5000
X Offset := 0.0000
Y Offset := 0.0000
Scan Date := 22, 06, 1999
Image # := 2
Image Type := ULTRASOUND
CASE # := 1
Patient name := BREAST1B
Scan type := TRANSVERSE
Pixel Offset := 0
Grid 1 Units := 0.0938
Grid 2 Units := 0.0938
Number Representation := UNSIGNED BYTE
Bytes per Pixel := 1
Number of Dimensions := 2
Size of Dimension 1 := 256
Size of Dimension 2 := 256
Z value := -3.0000
X Offset := 0.0000
Y Offset := 0.0000
and so on for the remainder of the scans.
Data are either in 16-bit, 2's complement, integer representation but are required to be within the 0 to 32767 range or in 8-bit unsigned byte within 0 to 255. Data is in raster order with the first pixel being the upper left of the image (i.e. the most negative x coordinate pixel and the most positive y coordinate pixel), the next pixel being just to the right of the first pixel until that raster line is complete, then all remaining raster lines until the last pixel (lower right).
Structures are connected sequences of three-dimensional coordinates which define volumes of interest such as the target volume. A "structure" has a variety of attributes, including a "name", "edition number", "color", free text "description", etc.
The organization of the points is that they are grouped together in planes which coincide with planes on which CT scans are centered. A given structure does not have to be defined in all planes in which scans exist, but the planes in which it is defined are contiguous. That is, no planes are "skipped".
Within a given plane, a structure will consist of one or more "segments" (usually just one). Each segment is a sequence of at least four (4) points which are connected and the last and first points must be the same (that is, the segment is "closed"). These points define a generally irregular curve which lies on the surface of the volume being defined. All segments need not have the same number of points. Segments in contiguous scans are assumed to be connected in some way so as to form the surface of the volume. The reason for permitting more than one segment per plane is so that Y-shaped or O-shaped structures may be defined.
The current definition of structures is tied closely to a scan sequence, paralleling what is currently done in most programs. More general definitions, requiring a more general data structure, may be needed in future. The keyword/value sequence "Structure format:=Scan-based" shall be included to permit subsequent expansion.
The following figure suggests how the components of a structure are arranged. The coordinates are in centimeters and are relative to the PATIENT COORDINATE SYSTEM defined above (Figure 7.1).
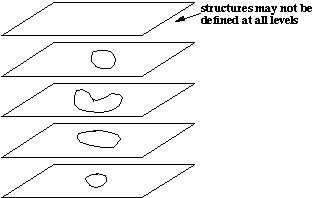
Figure 7.1
The data storage in a structure's image is "defined" through the example in Section 7.3. The data are placed in the buffer in the following order:
Number of levels (total # of scans)
Scan number (=1 for first scan, etc.)
Number of segments in this level (scan)
number of points in first segment
triplets of (x, y, z) coordinates, one per point, last=first
number of points in second segment
triplets of (x, y ,z) coordinates, one per point, last=first
Scan number (=2 for second scan,) Number of segments in this level (scan) number of points in first segment triplets of (x, y, z) coordinates, one per point, last=first number of points in second segment triplets of (x, y ,z) coordinates, one per point, last=first
Comments may be embedded in the data file if enclosed in quotes as documented in 3.2.1.
Scans must be contiguous on tape. This supports the data structure of structures which presumes that sequential contours are associated with sequential (contiguous) scans ordered monotonically with increasing value of the associated z coordinate. All scans must be referenced (in order) even if the structure does not exist in a particular slice. In this case the only data in the file will be the Scan # and the Number of Segments (0). See Section 7.3 for an example of this.
Required Keywords
Image # := actual image (file) number (see 4.4)
Image type := STRUCTURE
Case # := 1 for first patient,
2 for second patient, etc.
Patient name := patient identifier
Structure name := structure identifier (liver, heart,
etc.)
Number Representation := CHARACTER
Structure format := SCAN-BASED
Number of scans := same as # CT scans in the exchange
file set
Optional Keywords
Maximum # scans := 100 or system limit (may be set to
Number of scans value)
Maximum points per segment := 200 or system limit
Maximum segments per scan := 2 or system limit
Unit # := unit number or ID
Writer := person responsible for this data
Date written := AAPM format date (DD, MM, YYYY)
Structure edition := 1 or higher
Structure color := RED, GREEN, BLUE, YELLOW, MAGENTA,
CYAN OR WHITE
Structure description := Free form text
Study # of origin := for submitting institution's
identification
Orientation of structure := TRANSVERSE
Format of data in the image:
ASCII Text
Image # := 56
Image Type := STRUCTURE
Case # := 1
Patient Name := BREAST1B
Structure Name := EXTERNAL
Number Representation := CHARACTER
Structure Format := SCAN-BASED
Number of Scans := 55
Maximum # scans := 128
Maximum Points per Segment := 200
Maximum Segments per Scan := 4
Image # := 57
Image Type := STRUCTURE
Case # := 1
Patient Name := BREAST1B
Structure Name := TARGET
Number Representation := CHARACTER
Structure Format := SCAN-BASED
Number of Scans := 55
Maximum # scans := 128
Maximum Points per Segment := 200
Maximum Segments per Scan := 4
"NUMBER OF LEVELS" 55
"SCAN # " 1
"# OF SEGMENTS " 0
"SCAN # " 2
"# OF SEGMENTS " 0
"SCAN # " 3
"# OF SEGMENTS " 0
"SCAN # " 4
"# OF SEGMENTS " 0
(8 structure scan numbers omitted here)
"SCAN # " 13
"# OF SEGMENTS " 0
"SCAN # " 14
"# OF SEGMENTS " 0
"SCAN # " 15
"# OF SEGMENTS " 0
"SCAN # " 16
"# OF SEGMENTS " 0
"SCAN # " 17
"# OF SEGMENTS " 0
"SCAN # " 18
"# OF SEGMENTS " 1
"# OF POINTS " 15
-6.440, 5.850, -3.500
-6.230, 5.890, -3.500
(11 coordinate triplets omitted here)
-6.660, 5.620, -3.500
-6.440, 5.850, -3.500
"SCAN # " 19
"# OF SEGMENTS " 1
"# OF POINTS " 32
-6.260, 7.190, -3.000
-6.350, 7.240, -3.000
-6.350, 7.240, -3.000
(28 coordinate triplets omitted here)
-6.260, 7.190, -3.000
"SCAN # " 20
"# OF SEGMENTS " 1
"# OF POINTS " 27
-7.590, 7.580, -2.500
-7.300, 7.690, -2.500
etc.
Beam geometry's are to be be transferred as one data file per beam with the data file containing the information defining the beam aperture information. Some of the formalism herein is borrowed from the Foundation Library Specification and Virtual Machine Platform (VMP) Specification document from the Radiotherapy Treatment Planning Tools Collaborative Working Group (Tech. Report 91-1, Ira Kalet, Ph.D., Radiation Oncology Department RC-08, University of Washington, Seattle, WA 98125, USA).
There are several pieces of information required to be able to build a "treatment plan" using beam geometries. The first is the particular beam definition itself, including the prescribed dose per treatment of this field as well as the number of treatments delivered. Second is the identification of other beams that are treated in the same fraction(s) with this beam so that fractionation information may be obtained. Additionally, the grouping of all beams which are treated (or may be treated) is also provided so that a composite of all treatments may be reconstructed and the fractionation data with it.
The origin of the beam coordinate system (for the aperture definition) is defined with the treatment machine's collimator rotated to the neutral position (e.g. new Varian machines allow collimator angles from 90 to 270 degrees with 180 being the "neutral" position) and the gantry angle set such that the beam is pointed at the floor (down). The +y axis is toward the machine gantry when viewing along the beam's central axis with the gantry toward the top of your head. The +x axis is to your right when using the same view. All coordinates for apertures are in this unrotated coordinate system. All collimator, gantry and couch angles are defined to be zero for the gantry pointed down, the couch longitudinal axis orthogonal to the plane of gantry rotation and the collimator's +y axis is along the couch's longitudinal axis and is pointed toward the gantry. See Figure 8.1.
Angles are positive in the counter-clockwise (CCW) direction. CCW is defined from the above view for collimator and couch rotation and as viewed when looking into the gantry from the couch for the gantry rotation. The assumed patient orientation is with head to gantry. If the patient is being treated with foot to gantry, the keyword HEAD I/OUT must be used with a key value of OUT. The HEAD IN/OUT keyword may also be standardly used for head in as well but is required for head out treatments. For example, a right lateral beam for a patient oriented with head to gantry will have a gantry angle of 90 degrees, while the gantry angle would be 270 degrees for a right lateral beam with the patient's feet toward the gantry.
Beam shapes may be specified by MLC settings, contours for custom portal blocks and, for use with Peregrine and similar systems, by transmission maps. For a simple block, or MLC field, the map points inside the open regions of the beam would have a transmission value of 1.000. The map points under the MLC leafs or block will have transmission values appropriate with recommendations and/or requirements of receiving system. The 3D QA Center does not support the use of transmission maps for block specification.
Note that dynamic, conformal therapy and intensity modulation are not explicitly accounted for here and are left for future expansion.
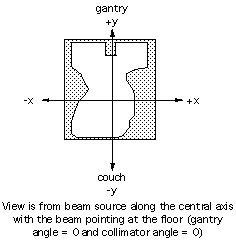
Figure 8.1
The data in the image file is as follows:
ADD3: For asymmetric collimator specifications the jaw which normally resides to the left (negative X in beam coordinates) or to the bottom (negative Y in beam coordinates) is specified first followed by the opposing jaw position. Again, note that a negative coordinate for an asymmetric jaw value implies that it has crossed the central ray. For instance, an asymmetric collimators setting of 11.0, 14.0 for X and -2.0, 8.0 for Y results in a 25.0 cm wide by 6.0 cm long rectangle which is centered at +1.5 cm in X and +5.0 cm in Y.
For APERTURE TYPE := COLLIMATOR
For APERTURE TYPE := BLOCK
For APERTURE TYPE := MLC_X or MLC_Y
For APERTURE TYPE := MLC_XY
For APERTURE TYPE := TRANSMISSION_MAP
If blocks are used, only one aperture definition is allowed although there is no strict limit on block definitions. This is to prevent system dependent ambiguity which would arise in the case of multiple apertures. The assumption this specification makes is that once a ray from a beam is blocked, it stays blocked. In the case of an aperture, all points outside of the contour are implicitly blocked, therefore they remain blocked.
Transmission Map Description
The transmission map specification involves three primary bits of data. The first is the matrix specification for the map for a rectangular matrix of square transmission elements. This specification includes the size of the square elements, the number of elements in each row and column and the coordinate of the center of the elements (not a corner). Another is a transmission value for the rectangular matrix made up of square elements where the transmission numbers represent the appropriate transmission value for the block material used according to the requirements of the receiving system. Points not under any block material will have a transmission value of 1.00 with lesser values for points under attenuators (MLC or block). Lastly, a map of block material thickness versus transmission value to define the physical characteristics of the portal shaping device. This implies that there are only as many distinct transmission values as are defined in the list of thicknesses and transmission values.
The order of the data in the file (for the transmission map) is identical to that used for compensating filters. The transmission values are specified in raster order from the most negative X and most positive Y coordinate (in beam coordinates) to the most positive X coordinate and most positive Y coordinate for the first row, followed by each subsequent row (see Figure 8.2).
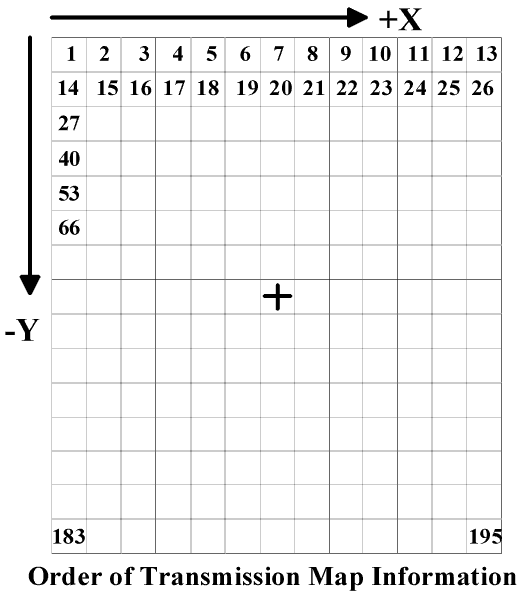
Figure 8.2
An example of the data in a transmission map beam data file is as follows. Note that the isocenter and collimator information is first in the file, followed by the transmission map followed by any compensator information.
"# of X Elements" 101, "# of Y elements" 85
"Size of square matrix element (cm)" 0.15
"Center of X1, Y1 (cm)" -7.5, 6.3
"# of transmission value thickness pairs" 2
"Pair #1" 1.0000, 0.0
"Pair #2" 0.0325, 8.1
"NX" 8, "NY" 6
"ROW #1"0.0325, 0.0325, 0.0325, 0.0325, 0.0325, 0.0325, 0.0325, (etc)
"ROW #2"0.0325, 0.0325, 0.0325, 1.0000, 1.0000, 1.0000, 1.0000, (etc)
Compensating filter information follows.
ADD4:Compensating Filters
Compensating filters may be specified in an abbreviated or extended form. The abbreviated form is identical to that used for Version 3.22 of this Specification. That uses only a "flag" to indicate that a compensating filter was used through use of the COMPENSATOR keyword and the appropriate keyvalue (NONE, 1D-X, 1D-Y, 2D, or 3D).
The extended form allows for either construction or attenuation information to be provided using the new keyword COMPENSATOR FORMAT. The key values available for use with this keyword are: THICKNESS, ATTENUATION, TISSUE, or NONE. Using the NONE keyvalue is identical to the results obtrained through using only the COMPENSATOR keyword with the appropriate flag and not including the COMPENSATOR FORMAT keyword (identical to the Version 3.22 capability). The other key values (THICKNESS, ATTENUATION and TISSUE) indicate that a matrix of compensating filter construction is being supplied in the beam data file. This matrix specification and data is in the data file following all other beam geometry information (isocenter, collimators, blocks and/or MLC specifications). In the case of ATTENUATION, the matrix values are fractional transmission (i.e. 0.25 indicates that 25% of the impinging radiation is transmitted). There is no explicit or implict statement about whether the attenuation values are narrow or broad beam. The matrix values in the data file for THICKNESS indicate the thickness of the construction material in cm. It is assumed that the receiving system has predefined information necessary to appropriately use this information for dose calculation (e.g. construction material). For TISSUE specified compensators, the matrix values correspond to the thickness of unit density tissue which must be accounted for. This generic specification may allow for appropriate interpretation by construction systems or devices.
2D or 3D Compensator Construction Specification
As the only difference between 2D and 3D compensators is the inclusion, or exclusion, of heterogeneity corrections for their design, they are specified in identical fashion as a two-dimensional grid defined at the NOMINAL ISOCENTER DISTANCE specified for the beam in which the delta-x between all columns in the matrix is uniform as is the delta-y between rows, but where the delta-x and delta-y are not required to be equal to each other (but, probably will be). The compensator matrix data is specified in raster order such that the starting coordinate specified is to the upper left (least X and greatest Y matrix element) of the grid (similar to the order of dose matrix values in a transverse plane). Because it is assumed that each matrix element occupies space, the starting coordinate specified (and the coordinates for other elements computed) are in the center of a region of attenuation with width delta-X and length delta-Y. Specifying the center of the matrix element causes the X1, Y1 coordinates to be offset by one-half the delta of the axis from the corner of the physical compensator (toward positive X and negative Y). The data is formatted as follows:
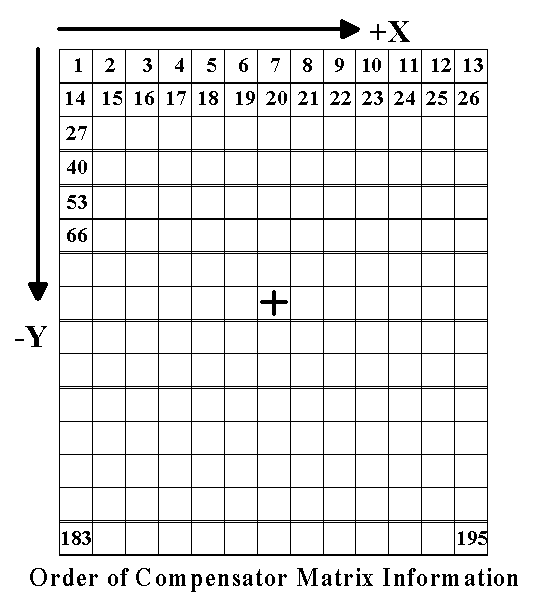
Figure 8.3
Figure 8.3 demonstrates the order of compensating filter data in the data file using the numbers in the individual compensator cells. Note that this is in raster order with a positive delta-X and a negative delta-Y. This figure shows the central ray of the beam through the center of a grid element, however, this is not required and the grid may align in any fashion with the major axes of the beam.
A simple (non-realistic) example of a 2D or 3D compensator construction text file follows. This sample is for a VERY SIMPLE compensator for a SMALL field for a beam with a NOMINAL ISOCENTER DISTANCE of 100.0 cm and for which the compensator matrix elements project to 1.5 cm wide at this distance. The collimator settings are symmetric along both axes and result in a field size of 10.0 cm x 7.0 cm at this same distance. The COMPENSATOR FORMAT is ATTENUATION. For compensating filter specification in thicknesses, it is assumed that the receiving system has some predefined understanding of the material used for construction. This compensator information follows the isocenter, collimator and blocking specification information.
"NX" 8, "NY" 6 "delta-X (cm)" 1.50, "delta-Y (cm)" 1.50 "X1, Y1" -7.25, 3.75 "Attenuation value per cm" 1.00 "ROW #1"0.872, 0.880, 0.820, 0.820, 0.850, 0.850, 0.900, 0.900 "ROW #2"0.900, 0.900, 0.820, 0.850, 0.850, 0.850, 0.900, 0.900 "ROW #3"0.900, 0.900, 0.850, 0.850, 0.850, 0.950, 0.950, 0.872 "ROW #4"0.900, 0.900, 0.850, 0.800, 0.900, 1.000, 0.950, 0.872 "ROW #5"0.872, 0.900, 0.850, 0.800, 0.850, 0.950, 0.900, 0.900 "ROW #6"0.872, 0.880, 0.820, 0.820, 0.850, 0.850, 0.900, 0.900
The 1D compensator (or custom step-wedge) is more simply specified as it contains only a single array corresponding to the axis across the steps (X or Y). Because the steps of these types of systems are not necessarily regularly spaced, the compensator is specified much like a cumulative histogram plot with each step being specified by a starting beam coordinate (at the NOMINAL ISOCENTER DISTANCE) and a thickness or attenuation value which is considered constant to the coordinate of the next step specified. Note that the type (ATTENUATION, TISSUE or THICKNESS) are handled in the same manner as that for 2D and 3D compensators. The slabs must be specified order of increasing coordinate (X or Y, as appropriate).
A simple example of a 1D compensator data file entry for a 1D-X compensator specified by THICKNESS follows:
"NX" 8 "Attenuation value per cm" 0.967 "SLAB #1 X-coordinate"-10.00, 0.000 "SLAB #2 X-coordinate" -9.00, 0.600 "SLAB #3 X-coordinate" -7.00, 1.200 "SLAB #4 X-coordinate" -3.00, 1.800 "SLAB #5 X-coordinate" 0.00, 2.400 "SLAB #6 X-coordinate" 1.00, 3.000 "SLAB #7 X-coordinate" 3.00, 3.600 "SLAB #8 X-coordinate" 6.00, 4.200 "SLAB #9 X-coordinate" 8.00, 4.800 "SLAB #10 X-coordinate" 10.00, 4.200 "SLAB #11 X-coordinate" 11.00, 0.000
There is no extrapolation or extension of compensator information beyond the coordinate values covered by the explicit compensator specification. Specifying a compensator smaller that the open field dimensions on the skin will have indeterminate results.
Following are the keywords for the Beam Geometry definition in the directory file:
Required Keywords
Image # := actual image (file) number (see 4.4)
Image Type := BEAM GEOMETRY
Case # := 1 for first case, 2 for second case
in file set, etc.
Patient Name := patient identifier
Beam # := Beam number in plan of origin (to
index with dose files later)
Beam Modality := X-RAY, ELECTRON, PROTON, NEUTRON, OTHER
Beam Energy(MeV) := Beam energy in MeV
Beam Description := Text Description of beam (i.e. LPO,
AP Boost, etc.)
Rx Dose Per Tx (Gy) := ICRU Reference point dose per treatment
(generally, isocenter dose)
Number of Tx := Number of treatments using this field
Fraction Group ID := ID to group beams of common fraction
Beam Type := STATIC, ARC
Collimator Type := SYMMETRIC, ASYMMETRIC, ASYMMETRIC_X,
ASYMMETRIC_Y
Aperture Type := BLOCK, MLC_X, MLC_Y, MLC_XY, COLLIMATOR, or
TRANSMISSION MAP
Collimator Angle := Collimator angle in degrees
Gantry Angle := Gantry angle in degrees (also start
angle for an arc beam)
Couch Angle := Couch angle in degrees
Nominal Isocenter Dist := Rotational source-isocenter distance
in cm or nominal treatment distance
(i.e. 80.0 cm for Co-60)
Number Representation := CHARACTER
Optional Keywords
Plan ID of Origin := Plan ID of beam origin for grouping
beams and doses
Aperture Description := Description of beam aperture
Aperture ID := Identifier of Aperture for beam
Wedge Angle := Wedge angle in degrees (required if
wedges are used for this beam)
Wedge Rotation Angle := 0, 90, 180, 270 ( required if wedges
are used for this beam) where:
0 - toe of wedge points toward +y beam axis
90 - toe of wedge points toward +x beam axis
180 - toe of wedge points toward -y beam axis
270 - toe of wedge points toward -x beam axis
Arc Angle := Arc angle in degrees (Req'd of ARC
Beam Type) it's sign should reflect the
stopping gantry angle.
Machine ID := text string uniquely identifying machine
parameter set used for dose calculation
Beam Weight := numeric value specifying beam weight used
(or to be used) for dose calculation with
definition of this value driven by the
WEIGHT UNITS keyword
Weight Units := MU, RELATIVE or PERCENT
MU is actual monitor unit (or time) setting
used for each treatment
RELATIVE is the fractional amount of total
beam on time for this beam versus the total
beam on time
PERCENT is the percentage amount of total
beam on time for this beam versus the total
beam on time
BEAM WEIGHT and BEAM UNITS are both required
if either one of them is used
Compensator := NONE, 1D-X, 1D-Y, 2D, 3D where:
1D is a customized step wedge along
specified beam axis
2D is a topographic correcting compensator
(an Ellis type for instance)
3D corrects for topography and heterogeneity
Compensator Format := THICKNESS, TRANSMISSION, TISSUE or NONE where:
THICKNESS indicates the compensator is
specified in ray thicknesses in cm
TRANSMISSION indicates the compensator is
specified in ray transmission values
TISSUE indicates the compensator is
specified in ray thicknesses in cm of tissue
NONE indicates the compensator's
construction is not specified (default if this
keyword not used for a compensator)
Head In/Out := IN, OUT where:
IN specifies this beam treated with
the patient's head toward the gantry
(prior to any couch rotation), and
OUT specifies this beam treated with
the patient's head away from the gantry
(prior to any couch rotation).
NOTE: Orientation is assumed to be
head in unless otherwise specified.
This keyword is required for a foot
in treatment.
Format of data in the image file:
ASCII TEXT
Image # := 25
Image Type := BEAM GEOMETRY
Case # := 1
Patient Name := Joe Smith
Beam # := 1
Beam Modality := X-RAY
Beam Energy(MeV) := 18
Beam Description := AP Port
Rx Dose Per Tx (Gy) := 1.00
Number of Tx := 25
Beam Type := STATIC
Plan ID of Origin := final
Collimator Type := ASYMMETRIC_X
Aperture Type := BLOCK
Aperture Description := AP Portal Large Field
Collimator Angle := 0
Gantry Angle := 0
Couch Angle := 0
Nominal Isocenter Dist := 100.0
Aperture ID := AP Port Block
Compensator := 1D-Y
Number Representation := CHARACTER
Fraction Group ID: := 1
Head In/Out: := IN
"Isocenter coordinate" 1.0, -2.5, 15.2
"Collimator Setting x" 11.0, -2.5
"Collimator Setting y" 15.0
"# of block contours" 2
"Block #1 type contour encloses open portal" 0
"Transmission under block" 0.03125
"# of block coordinate pairs" 6
-10.5, 7.0, -3.0, 7.0, -3.0, -7.2, -5.0, -4.3, -9.5, -6.5
-10.5, 7.0
"Block #2 type contour encloses spinal shield" 1
"Transmission under block" 0.03125
"# of block coordinate pairs" 5
-7.5, 7.5, -5.5, 7.5, -5.5, -7.5, -7.5, -7.5, -7.5, 7.5
"Compensating filter data as shown above"
Here is a short example of a multi-leaf data file (MLC_X) with asymmetric collimators in x (ASYMMETRIC_X). All coordinates are defined at the Nominal Isocenter Distance. Note that words in quotes are to be ignored by the processing program as documented in Section 3.1.2.
"Isocenter coordinate" 1.0, -2.5, 15.2
"Collimator Setting x" 11.0, -2.5
"Collimator Setting y" 15.0
"Number of Leaf Pairs" 26
"Leaf center y positions" -12.5, -11.5, -10.5, -9.5, -8.5, -7.5
-6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5
3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5
"Leaf pair thickness" 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0
1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0
1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0
"Leaf extensions for Y1" -8.81, 8.81
"Leaf extensions for Y2" -8.81, 8.81
"Leaf extensions for Y3" -8.81, 8.81
"Leaf extensions for Y4" -8.81, 8.81
"Leaf extensions for Y5" -8.81, 8.81
"Leaf extensions for Y6" 6.86, 6.95
"Leaf extensions for Y7" 7.93, 7.96
"Leaf extensions for Y8" 8.31, 8.26
"Leaf extensions for Y9" 8.31, 8.25
"Leaf extensions for Y10" 8.30, 8.25
"Leaf extensions for Y11" 8.30, 8.25
"Leaf extensions for Y12" 8.30, 8.25
"Leaf extensions for Y13" 8.29, 8.24
"Leaf extensions for Y14" 8.29, 8.23
"Leaf extensions for Y15" 7.91, 7.79
"Leaf extensions for Y16" 7.50, 7.36
"Leaf extensions for Y17" 6.50, 6.92
"Leaf extensions for Y18" 6.68, 6.49
"Leaf extensions for Y19" 6.27, 6.05
"Leaf extensions for Y20" 5.86, 5.62
"Leaf extensions for Y21" 5.45, 5.18
"Leaf extensions for Y22" 5.04, 4.74
"Leaf extensions for Y23" 4.63, 4.31
"Leaf extensions for Y24" -8.81, 8.81
"Leaf extensions for Y25" -8.81, 8.81
"Leaf extensions for Y26" -8.81, 8.81
"Compensating filter data as shown above"
This image type supports the exchange of digitized simulation films, digitized portal films, on-line portal images, and computed images (i.e. DRR's). The basic information to be included is the pixel data itself and identifiers so that one image may be distinquished from another when multiple images of the same field are used. The pixels themselves are to be transferred in raster order where the first pixel is the upper left pixel of the image with the most rapid change in position with changing pixel is to the right of the image. The last pixel in the image is the lower right.
The film coordinate system is identical to that used for the Beam Geometry images with respect to the x and y offsets and axes. The DRR digital film image is assumed to be aligned with the unrotated collimator. For example, if the pixel image were to be displayed on a monitor with the collimators superimposed, the collimator edges would be rotated (relative to the edges of the display) if the collimator angle is other than 0 degrees (or a multiple of 90 degrees). If DRR's are aligned with the collimator edges, regardless of the collimator rotation, the COLLIMATOR ANGLE keyword must be used and its' value must be the collimator angle for the associated beam. This angle will be assumed to be zero (implying that the film does not rotate with the collimator) unless this keyword and appropriate value are used.
There are parameters which may be included in the directory to describe a digital film which are designed to define the alignment of the image in the associated radiation beam. While these parameters are necessary for any digital film image (particularly for DRR's), which does not have either a fiducial grid or a port outline on it from which such alignment may be derived, they are not generally required for SIMULATOR or PORT image files. Generally, since this alignment information is available for DRR images, such alignment data is required. The affected keywords are: Grid 1 Units, Grid 2 Units, Source Image Distance, X offset, Y offset and Collimator Angle. Where zero (0) is implicit in the image data (for instance, DRR's are generally constructed such that the central ray is in the geometric center of the pixel image) these keywords are not required. For DRR images Grid 1 Units, Grid 2 Units, Source Image Distance are required keywords, while the use of X offset, Y offset and Collimator Angle depend on the context of the image generation as described with the keyword. None of these keywords is required for SIMULATOR or PORT images.
The pixel data is transferred in a fashion similar to the CT pixels, in that they may be 16 bit unsigned integer values whose range is restricted to 0 to 32767 or may be in a range of 0 to 255 for unsigned byte data. The number of bits per pixel acutally containing data may be specified in order to facilitate the use of local packing and display software.
Since it is possible to have multiple images of the same port in one day, the combination of date and film number uniquely identify a film. Generally, the film number will be 1, but multiple images of the same port in a day are supported through this method.
ADD5: In order to facilitate the exchange of digital film images without having an attached beam in a fraction group (for instance a urethrogram film or perhaps orthogonal isocenter verification films without corresponding beams in the treated fraction groups), the BEAM # and BEAM DESCRIPTION keywords have been made optional. The condition to their optional nature is that if they are not used, the FILM DESCRIPTION keyword must be used and vice versa.
Required Keywords
Image # := actual image (file) number (see 4.4)
Image Type := DIGITAL FILM
Case # := 1 for first case, 2 for second case in
file set
Patient Name := Patient Identifier
Film Number := Number of film on particular date (i.e.
1, 2, etc.)
Film Date := Date digital image acquired (DD, MM, YYYY)
Film Type := SIMULATOR, DRR, PORT
Number of Dimensions := 2 (always)
Size of Dimension 1 := number of rows
Size of Dimension 2 := number of cols
Number Representation := TWO'S COMPLEMENT INTEGER (for 2 bytes
per pixel) or UNSIGNED BYTE (for 1 byte
per pixel)
Bytes per Pixel := 1 or 2 (must index with Number
Representation)
Optional Keywords
Beam # := Beam number in plan of origin (to tie
image with) Required if film belongs to a
beam in a submitted fraction group.
Beam Description := Text description of beam generating image
Required if film belongs to a beam in a
submitted fraction group
Film Description := Text Description of film
Required if BEAM # and BEAM DESCRIPTION
keywords not used and must be the same
identical string for all appropriate films
(i.e. AP ISOCENTER, RT LAT ISOCENTER, etc.)
Grid 1 Units := pixel width (cm) (required for DRR's)
Grid 2 Units := pixel length (cm)(required for DRR's)
Source Image Distance := equivalent to TFD (cm)(required for DRR's)
X Offset := X offset from geometric center of image
to central ray of the beam (required
for DRR's where central ray is not
in geometric center of pixel image)
Y Offset := Y offset from geometric center of image
to central ray of the beam (required
for DRR's where central ray is not
in geometric center of pixel image)
Film Source := FILM, ONLINE, COMPUTED
Unit Number := Unit number film image acquired from
OD Scale := Scale factor to convert pixel values
to optical density
Bits per Pixel := number of bits actually used for pixel
information
Collimator Angle := collimator angle in degrees (reflects
the collimator angle for the associated
beam) if the edges of the image are
parallel to the collimator edges.
This is required only for DRR's which
are aligned with the collimator edges
and which do not have the portal outline
superimposed on the DRR image. It is
not required for DRR's which are aligned
with the unrotated collimator or for
digitized films or on-line images
(SIMULATOR and/or PORT images).
Format of data in the image file:
Binary Data
Image # := 37
Image Type := DIGITAL FILM
Case # := 1
Patient Name := Joe Smith
Beam # := 6
Beam Description := Left Lateral Beam
Film Date := 15,11,1993
Film Number := 1
Film Type := SIMULATOR
Number of Dimensions := 2
Size of Dimension 1 := 480
Size of Dimension 2 := 512
Grid 1 Units := 0.215
Grid 2 Units := 0.200
Source Image Distance := 140.0
X Offset := 0.0
Y Offset := 2.3
Number Representation := TWO'S COMPLEMENT INTEGER
Bytes per Pixel := 2
Film Description := verification simulation film
Film Source := FILM
Image # := 38
Image Type := DIGITAL FILM
Case # := 1
Patient Name := Joe Smith
Beam # := 6
Beam Description := Right Lateral Beam
Film Date := 15,11,1993
Film Number := 2
Film Type := SIMULATOR
Number of Dimensions := 2
Size of Dimension 1 := 480
Size of Dimension 2 := 512
Grid 1 Units := 0.215
Grid 2 Units := 0.200
Source Image Distance := 140.0
X Offset := 0.0
Y Offset := 2.3
Number Representation := UNSIGNED BYTE
Bytes per Pixel := 1
Film Description := first day port image
Film Source := ONLINE
Image # := 39
Image Type := DIGITAL FILM
Case # := 1
Patient Name := Joe Smith
Film Description := AP Isocenter
Film Date := 15,11,1993
Film Number := 1
Film Type := SIMULATOR
Number of Dimensions := 2
Size of Dimension 1 := 640
Size of Dimension 2 := 752
Number Representation := UNSIGNED BYTE
Bytes per Pixel := 1
Film Source := FILM
Data may be in 16-bit, 2's complement, integer representation wherein the 2's complement is never really used as the values are required to be in the range of 0 to 32767. The pixel data may also be in unsigned byte data in which case the pixel values are between 0 and 255. Data is in raster order with the first pixel being the upper left-hand pixel in the image.
A dose distribution is the result of a calculation of dose at one or more points throughout the patient, for a particular configuration of beams - that is, for a particular "plan". Although in general, one might calculate doses on a completely irregular grid of points this is rarely done in practice and the proposed format is for a fairly regular grid, namely one in which a two dimensional array of points is defined in one or more parallel planes. This format naturally accommodates the computation of doses on a 2-D array of points in each CT scan, and recognizes that such scans may not be at equally spaced intervals. It permits the transfer of dose calculations throughout a volume, or in a single plane - or, indeed, along a line or at a single point. Planes may be other than parallel with the scan sections however, thus supporting calculations in sagittal or coronal planes. At present planes oblique to the major axes of the scans, or arbitrarily located points of calculation are not supported.
The points at which the doses are defined are assigned coordinates within the Patient Coordinate System. We first describe the coordinate definitions for the case of arrays defined in planes parallel to transverse sections (i.e. CT scans), and then indicate some differences when the planes are sagittal or coronal. The number of planes (>=1) and a list of the z-values is specified. Within a plane a rectangular array of points is defined by specifying the x, y coordinates of the upper left hand corner point (as viewed from the patient's feet), the x and y increments per point, and the number of points along the x-axis and along the y-axis. The z values for each plane may be unequally spaced and are therefor individually specified. For transverse planes these z values would normally be identical to those of some or all of the CT sections, but this is not required. The order of the planes should be that of increasing value of z.
To preserve the integrity of the right-handed cartesian coordinate system, some sign conventions must be obeyed when sagittal or coronal planes are used. The coordinates for single planes as presented to the observer are as follows:
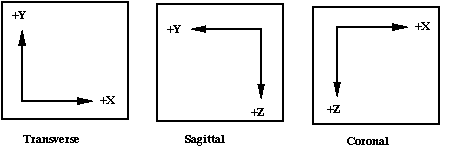
Figure 10.1
These sign conventions have implications for the various parameters as follows:
| PARAMETER | TRANS. | SAG. | COR. |
|---|---|---|---|
| (Horiz, vert) coords of points | x, y | z, y | x, z |
| Usual signs of coords of ULH corner | -, + | +, + | -, - |
| Usual sign of horizontal increment | + | - | + |
| Usual sign of vertical increment | - | - | + |
| Coordinate associated with plane change | z | x | y |
Note that these conventions need not be obeyed in the definition of pixel size of CT scans. The vertical size is permitted to be positive for CT scans to conform to conventional usage and is interpreted as the absolute value of the pixel height, rather than a signed increment.
The units in which doses are given are up to the originator of the data. They must be in absolute dose units such as Gray. Relative and Percent are no longer supported in the Dose Units keyword and are now implicit by the inclusion of the Dose Scale keyword, where the Dose Scale keyword is used only if scaling is necessary. The dose values in the image file are multiplied by the Dose Scale value to obtain the Dose Units specified. A 1.00 is assumed for the Dose Scale value unless it is explicitly stated with the Dose Scale keyword.
Dose distributions other than Physical dose, such as of Effective dose, LET, OER or dose uncertainty, are supported through the use of the "Dose Type" keyword.
The Fraction Group ID allows multiple dose distributions to be submitted which will allow for fractionation information to be extracted for both targets and normal tissues and is used to tie Beam Geometry files to a particular dose file (multiple Beam Geometry files may point to the same DOSE file through the Fraction Group ID). All beams contributing dose to this distribution shall have an identical Fraction Group ID in their beam geometry specification.
The Plan ID of Origin is similar to the Fraction Group ID except that instead of being used to tie beam files to the dose file, it is used to tie Seed Geometry files to a Dose file. Unlike the Fraction Group ID, there can only be one Seed Geometry file which points at a given dose file (i.e. there is a one-to-one correspondence).
While both the Fraction Group ID and Plan ID of Origin keywords are listed as optional, when used for RTOG 3D CRT protocol patients, they are required as appropriate.
TEXT (ASCII) DOSE SPECIFICATION
The data storage in a dose image is "defined" through the example given in Section 10.3. The data are placed in the buffer in the following order:
Number of planes (e.g. 19)
Z-coordinate of first constant z plane (for e.g. z = -120.556)
A sequence of real numbers representing the dose at each grid point at this z value. X
value (dimension 1) varies faster:
0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000
4.641, 11.785, 12.031, 10.608, 10.324, 10.258, 10.202
10.139, 10.125, 10.125, 10.118, 0.000, 0.000, 10.117
10.132, 10.148, 10.145, 10.145, 10.151, 10.183, 10.234
Z-coordinate of second constant z plane (for e.g. z = -119.616)
A sequence of real numbers representing the dose at each grid point at this z value. X
value (dimension 1) varies faster:
0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000
2.011, 9.881, 11.476, 10.608, 10.324, 10.258, 10.202
10.139, 10.125, 10.125, 10.118, 0.000, 0.000, 10.117
Doses may also be conveyed in a more succinct, binary format. In order to facilitate this format several additional (otherwise optional) keywords must be specified. Doses using the binary format must meet the following requirements:
The optional keywords required for binary dose specification may not be usedwith text dose specification. The order of the dose matrix elements is identical to that used for the text representation excepting that the Z coordinate is no longer specified (nor is the plane count). As with all binary files, no text is supported in the file (e.g. comments in quotes).
Required Keywords
Image # := actual image (file) number (see 4.4)
Image Type := DOSE
Case # := 1 for first patient, 2 for second
patient, etc
Patient Name := patient identifier
Dose Units := GRAYS, RADS, CGYS
Orientation of Dose := TRANSVERSE
Number Representation := CHARACTER
Number of Dimensions := 3
Size of dimension 1 := # horizontal points (>=1)
Size of dimension 2 := # vertical points (>=1)
Size of dimension 3 := # of planes (>=1)
Coord 1 of first point := x coord (cm) for transverse, etc.
Coord 2 of first point := y coord (cm) for transverse, etc.
Horizontal grid interval := delta-x (cm) for transverse (>0)
Vertical grid interval := delta-y (cm) for transverse (<0)
Optional Keywords
Dose # := # identifying this distribution
Dose Type := PHYSICAL, EFFECTIVE, LET, OER, ERROR
Unit # :=
Writer :=
Date written := date (DD, MM, YYYY)
Dose description := free text
Dose edition :=
Plan # of origin :=
Plan edition of origin :=
Study # of origin :=
Version # of program := planning program identification
x coord of normalizn point := cm
y coord of normalizn point := cm
z coord of normalizn point := cm
Dose at normalizn point := should result in units specified
above after being multiplied by
the Dose Scale
Dose error := NOMINAL, MINIMUM, or MAXIMUM
(for dose range submissions)
Fraction Group ID := ID grouping beams of common
fraction for the doses in this
image file
Number of Tx := Number of times this fraction
(Fraction Group ID) treated to
achieve total doses in this file
Dose Scale := Scale factor to convert doses in
image file to absolute doses in
the units specified in the Dose Units.
(assumed to be 1.00 if not specified)
Coord 3 of first point := z coord (cm) for first transverse plane
Depth grid interval := delta-z (cm) between each subsequent
transverse dose plane (>0)
Plan ID of origin := Plan ID of SEED GEOMETRY to required to tie
DOSE file to SEED GEOMETRY file
All coordinates and differences are expressed in centimeters in the patient coordinate system.
Format of data in the image:
ASCII text.
Image # := 57
Image Type := DOSE
Case # := 1
Patient Name := CHEST1C
Dose # := 1
Dose Type := PHYSICAL
Dose Units := GRAYS
Orientation of Dose := TRANSVERSE
Number Representation := CHARACTER
Number of Dimensions := 3
Size of dimension 1 := 116
Size of dimension 2 := 74
Size of dimension 3 := 101
Coord 1 of first point := -19.3000
Coord 2 of first point := 14.3000
Horizontal grid interval := 0.3000
Vertical grid interval := -0.3000
Dose description := 4FLD CHESTWALL WITH BOLUS
Plan # of origin := 26
Fraction Group ID := 1
Number of Tx := 25
Dose Scale := 0.01
"Number of planes is " 101
"Z-coordinate is " -15.200
0.000, 0.000, 0.000, 0.000, 0.012, 0.012, 0.013, 0.013
0.014, 0.015, 0.016, 0.016, 0.017, 0.018, 0.019, 0.019
0.020, 0.021, 0.022, 0.022, 0.023, 0.024, 0.024, 0.025
0.026, 0.026, 0.027, 0.028, 0.028, 0.029, 0.029, 0.030
0.030, 0.031, 0.031, 0.032, 0.032, 0.032, 0.033, 0.033
0.033, 0.033, 0.033, 0.033, 0.033, 0.033, 0.033, 0.033
0.033, 0.032, 0.032, 0.032, 0.031, 0.031, 0.031, 0.030
. . . . . . . .
. . . . . . . .
. . . . . . . .
0.030, 0.029, 0.029, 0.028, 0.027, 0.027, 0.026, 0.026
0.025, 0.024, 0.024, 0.023, 0.022, 0.021, 0.021, 0.020
0.019, 0.018, 0.018, 0.017, 0.016, 0.015, 0.014, 0.014
"Z-coordinate is " -15.000
0.013, 0.013, 0.012, 0.012, 0.011, 0.011, 0.011, 0.010
0.010, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000
0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000
. . . . . . . .
. . . . . . . .
. . . . . . . .
0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000
0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000, 0.000
0.012, 0.012, 0.013, 0.013, 0.014, 0.015, 0.015, 0.016
0.017, 0.018, 0.019, 0.019, 0.020, 0.021, 0.022, 0.022
0.023, 0.024, 0.024, 0.025, 0.026, 0.026, 0.027, 0.027
ADD6: The data file for binary formatted dose data consists of two byte integer values restricted to the values from 0 to 32767 packed with the most significant byte first (identical to the numeric format used for CT scans) written in raster order for each axial dose plane. Each subsequent axial plane's dose values are required to be in order of increasing Z coordinate. Any padding required for buffering (for tape writing only) is required only after the last dose point of the last axial plane is written to the file.
Dose-volume histograms (DVH) provide a "pre-digestion" of the doses provided in a 3-D dose distribution with corresponding anatomic structures. While there are several different methods which may be used to display the DVH data, the underlying data is the same: A bin of dose range and a volume associated with the dose range. DVH's are transferred as one structure per image file.
The data in the image file itself is simply an array of doublets where the first value in the doublet is the lower end of the dose bin and the second value is the volume associated with the dose bin. The doses may be in either absolute dose or percent dose and may be converted back and forth using directory information. The volume may be in units of percent or of cubic centimeter (cc) and may be converted back and forth with the additional information available in the directory information for the image file. The dose bins are required to be uniformly spaced and included in the data file from zero dose to the highest dose for which any non-zero volume is identified and no gaps are allowed.
The scaling of relative or percent doses or volumes are performed by multiplying the relative dose or volume value by the appropriate scale value.
Required Keywords
Image # := actual image (file) number (see 4.4)
Image Type := DOSE VOLUME HISTOGRAM
Case # := 1 for first case, 2 for second case
in file set
Patient Name := Patient Identifier
Structure Name := name of structure
Dose Units := GRAYS, CGYS, RADS
Dose Type := ABSOLUTE, PERCENT, RELATIVE
Volume Type := ABSOLUTE, PERCENT, RELATIVE
Number of Pairs := Number of dose/volume pairs in image
file
Maximum # Pairs := Maximum number of dose/volume pairs
allowed
Number Representation := CHARACTER
Plan ID of Origin := ID of plan DVH's calculated from.
Indexes with beams and dose
distributions.
Optional Keywords
Dose Scale := Scales percent or relative dose to
absolute dose (Required if dose type
is not ABSOLUTE)
Volume Scale := Scales percent or relative volume
to cc's (Required if volume type is
not ABSOLUTE)
Date of DVH := Date DVH calculated (DD, MM, YYYY)
Format of data in the image file:
ASCII TEXT
Image # := 39
Image Type := DOSE VOLUME HISTOGRAM
Case # := 1
Patient Name := Joe Smith
Structure Name := Rectum
Plan ID of Origin := final
Dose Units := GRAYS
Dose Type := ABSOLUTE
Volume Type := RELATIVE
Volume Scale := 203.1
Number of Pairs := 100
Maximum # Pairs := 1001
Number Representation := CHARACTER
Date of DVH := 15,11,1993
Image # := 40
Image Type := DOSE VOLUME HISTOGRAM
Case # := 1
Patient Name := Joe Smith
Structure Name := PTV
Plan ID of Origin := final
Dose Units := GRAYS
Dose Type := ABSOLUTE
Volume Type := RELATIVE
Volume Scale := 203.1
Number of Pairs := 100
Maximum # Pairs := 1001
Number Representation := CHARACTER
Date of DVH := 15,11,1993
"Minimum Bin Dose, Fractional Volume"
0.00, 0.05
1.00, 0.00
2.00, 0.06
. . . .
. . . .
. . . .
100.00, 0.00
Note that the volume associated with each bin dose is that volume which explicitly falls into that dose bin (hence the zero volume values for 1.00 Gy above sandwiched between the 0.00 Gy and 2.00 Gy bins.
Seed geometry files are used to convey the geometric distribution of permanently implanted I125 or Pd103 seeds. These seed distributions may be indexed with an image data set (CT, MRI or Ultrasound), or may be independent of any image set. The information provided in this file should be adequate to calculate the dose distribution with minimal modification of the incoming data by the receiving institution. Multiple seed distributions are only supported in a single file set if they comprise the complete implant in which varying seed activities and/or types are used in the same implant.
NOTE: It is assumed that if any image files (CT/MR/US) are contained within the same digital file set, that the seed coordinates are consistent with the coordinates of the images (i.e. the seeds are registered with the image set). If there are no images with which the seed coordinates are registered, then no image files are allowed to be provided in the same digital file set. This is to simplify the specification of registered versus unregistered seed coordinates.
The fundamental information contained in the directory entries for a Seed Geometry file are:
The data file associated with the directory entries consists of only coordinate triplets (in cm) for each of the number of seeds specified in ASCII (text) format.
Required Keywords
Image # := actual image (file) number (see 4.4)
Image Type := SEED GEOMETRY
Case # := 1 (or registered case number)
Patient Name := Patient Identifier
Seed Model := model identifier or manufacturer of seed
Isotope := I125 or PD103
Seed Strength := value corresponding to strength units specified
Strength Units := MCI or CGYCM2PERHR
Date of Implant := date (DD, MM, YYYY)
Number of Seeds := Number of seeds in image file (implant)
Number Representation := CHARACTER (format of data in data file)
Plan ID of Origin := ID of plan seed distribution from
Indexes with dose distributions.
Format of data in the image file:
ASCII TEXT
Image # := 42
Image Type := SEED GEOMETRY
Case # := 1
Patient Name := Joe Smith
Seed Model := 6711
Isotope := I125
Seed Strength := 0.43
Strength Units := MCI
Date of Implant := 23, 06, 1999
Number of Seeds := 27
Number Representation := CHARACTER
Plan ID of Origin := Preplan
Image # := 44
Image Type := SEED GEOMETRY
Case # := 1
Patient Name := Joe Smith
Seed Model := Model 2
Isotope := I125
Seed Strength := 0.38
Strength Units := MCI
Date of Implant := 23, 06, 1999
Number of Seeds := 63
Number Representation := CHARACTER
Plan ID of Origin := Actual Plan
12.3 Example of Seed Geometry Image File
" X (cm), Y (cm), Z (cm)"
"Seed #1" 0.00, 0.05, 5.00
"Seed #2" 0.00, 0.05, 5.90
"Seed #3" 0.00, 0.05, 7.20
"Seed #4" 0.00, 0.05, 8.10
. . . .
. . . . (intervening 80 seeds not shown)
. . . .
"Seed #85" 3.00, 3.25, 4.70
Document maintained by Walter R. Bosch
Last modified: